|
The Sethuraman Lab, represented by Alex, Priyanshi, Raya, Tamsen and Trevor recently attended the 2024 SCalE Meeting, hosted by CalTech. Alex presented some of our recent research work on the genomic history of domestication in hops. Watch this space for our upcoming manuscript describing these results!
0 Comments
 Figure 1. Locations where sturgeon tissue samples were collected from presumed captive (red) and wild (purple) populations. Captive populations included three coastal fish markets in Batumi, Poti, and Tskaltsminda one inland fish market in Tbilisi, and an aquaculture facility. Wild populations were sampled from the Black Sea, Rioni River, and the mouth of the Rioni River. Our new collaborative study that examines genomic evidence for the presence of protected white sterlet, beluga, and stellate sturgeon in fish markets in Tbilisi, Georgia, often hybridizing with locally farmed sturgeons, led by Tamar Beridze, a visiting scholar in the Sethuraman lab in Fall 2022 is now published in Diversity! Read here.
P. quatuordecimpunctata and H. variegata invasive pop-gen study now published in Biological Control!5/8/2024 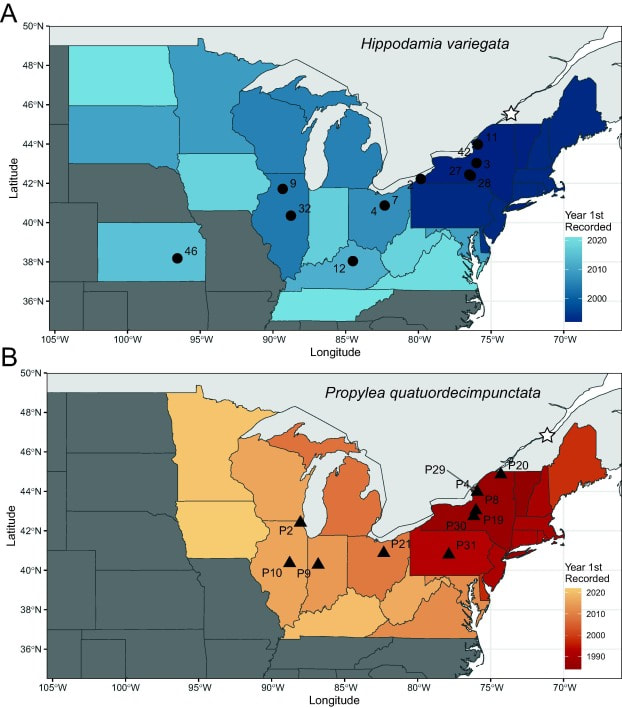 Fig. 1. Sampling of (A) H. variegata and (B) P. quatuordecimpunctata across their invasive United States ranges with numerical locality identifiers. Parenthetical numbers denote the number of sequenced samples retained for genetic analyses. Color gradient displays the year of first recorded specimen in each state. Star icon shows approximate location of North American introduction. Our new study on invasive population genomics and behavioral phenology in response to photoperiods in two species of lady beetles used in importation biological control - Propylea quatuordecimpunctata and Hippodamia variegata is now published in Biological Control! Read here.
Christopher Morrissey successfully defended his Masters thesis, and we couldn't be prouder of him! As part of his thesis, he developed an R package called MethylMapR, which can be used to characterize the functional methylome in prokaryotes - watch this space for our upcoming publication describing this package. Congratulations, Chris!
Graduate student Trevor Mugoya was recently awarded the 2024-2025 SDSU Presidential Graduate Research Fellowship! Congratulations, Trevor! We are so proud of you and look forward to your continued success at SDSU and beyond.
Our manuscript describing a high-quality genome of the convergent lady beetle, Hippodamia convergens is now out in G3! We used the Dovetail Genomics platform to generate long (PacBio) and short read (Illumina) data, benchmark assembly with wtdbg2, Flye and Canu; with a winning Canu+purge_dups+RagTag scaffolded assembly with an N50 of 89 Mbps with a majority of the genome in a handful of scaffolds. We also resolve its species tree among Coleopterans using whole genome phylogenetic reconstruction. Congratulations to Gavrila Ang and Andrew Zhang on this neat study! Read here.
Loggerhead shrike pop-gen + geometric morphometrics manuscript published in Ecology and Evolution!3/19/2024 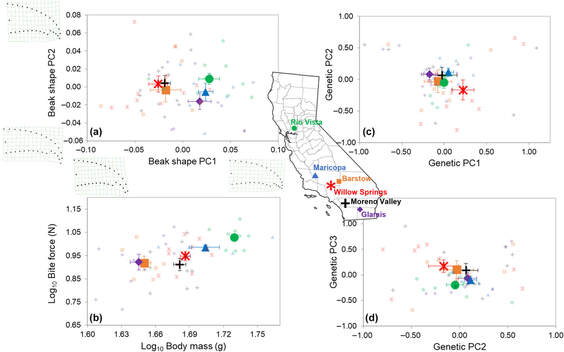 Location means ± SE along bivariate phenotypic (left side) and genetic (right side) spaces: (a) The first two principal component axes of upper beak shape, including shape deformation grids showing shape differences along the extremes of each axis; (b) log10 bite force vs. log10 body mass; (c) the first two axes of the genetic principal component analysis based on microsatellite data; (d) the second and third axes of the genetic PCA. Symbol color and shape combinations correspond to different sampling locations throughout California labeled on the map (individual data points are semi-transparent). Our manuscript that describes phenotypic differences despite the presence of little population structure and lots of gene flow between Californian populations of the loggerhead shrike, Lanius ludovicianus is now published in Ecology and Evolution! Congratulations to Sethuraman Lab alum, Gwen Wulf on this publication! Read the paper here.
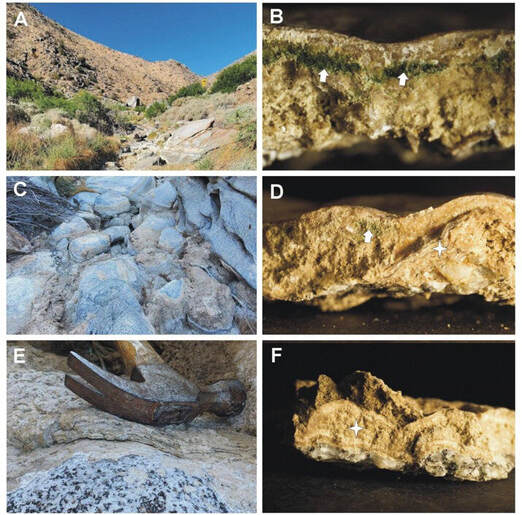 The ephemeral San Felipe Creek in Anza Borrego Desert (A). Samples were collected from the surface of boulders found in the bed of the desiccated creek (C and E). Samples imaged under a dissecting scope (B, D, and F) show characteristic features of stromatolites including endolithic cyanobacteria underneath a layer of calcite (arrows in panels B and D) and multiple laminate layers of organic and inorganic materials (asterisks in panels D and F). DNA was isolated from the stromatolites shown in panels B and E. Our article describing the microbial metagenome of calcareous stromatolite formations in the Anza Borrego desert in California, led by talented NSF REU scholars from Summer 2022/2023 has now been published in the American Society for Microbiology's Resource Announcements. Read the article here!
The Sethuraman Lab recently presented our research at the TAGC24 meeting in Washington D.C., with PhD student Margaret Wanjiku giving a talk on her first thesis chapter on a series of statistical tests to detect and account for gene flow from archaic ghost populations, PhD student Alexandra McElwee-Adame presenting a poster on her first thesis chapter on the evolutionary genomics of domestication in hops (Humulus lupulus L.), MS candidate Priyanshi Shah presenting her work on telomere length variation in diverse human populations, and Undergraduate student Scott Monahan presenting work on heritability and plasticity in body size of Dinocampus coccinellae, a parthenogenetic wasp. We also had a grand time being tourists around D.C.! Cheers to more fun science meetings in the future! Dr. Sethuraman was recently awarded the 2024 SDSU Research, Scholarship, & Creative Activities Mentor Award, which was presented by Dr. Hala Madanat, Vice President for Research & Innovation at the S3 symposium.
|
Arun Sethuraman
|





 RSS Feed
RSS Feed