I develop and maintain methods (often collaboratively) for population genetic/genomic analyses, several of which are accessible on my GitHub page. Here are descriptions of some of these methods, and links to download and use them. Feel free to contact me with bug reports/questions.
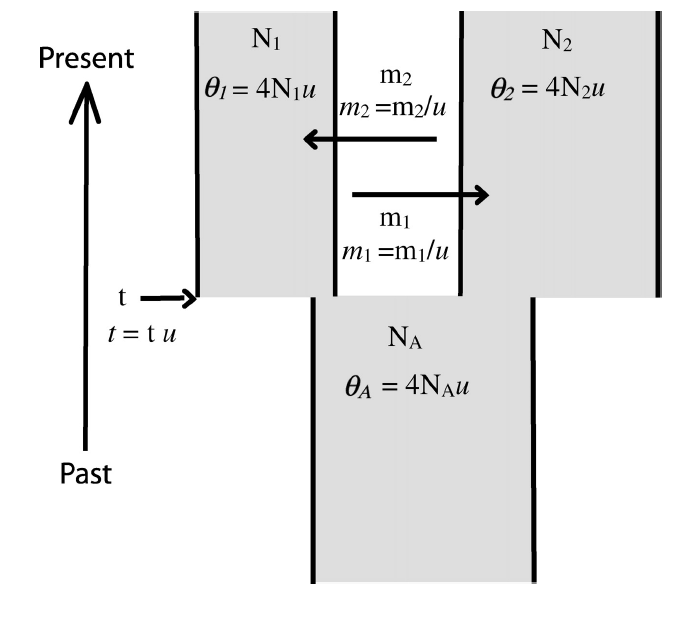
IMa2pParallelized Isolation with Migration (IM) analyses of haplotypic data to estimate evolutionary demography (effective population sizes, migration rates, and divergence times) of populations. Read our paper on IMa2p, published in Molecular Ecology Resources here. Code is available on my GitHub page here.
|
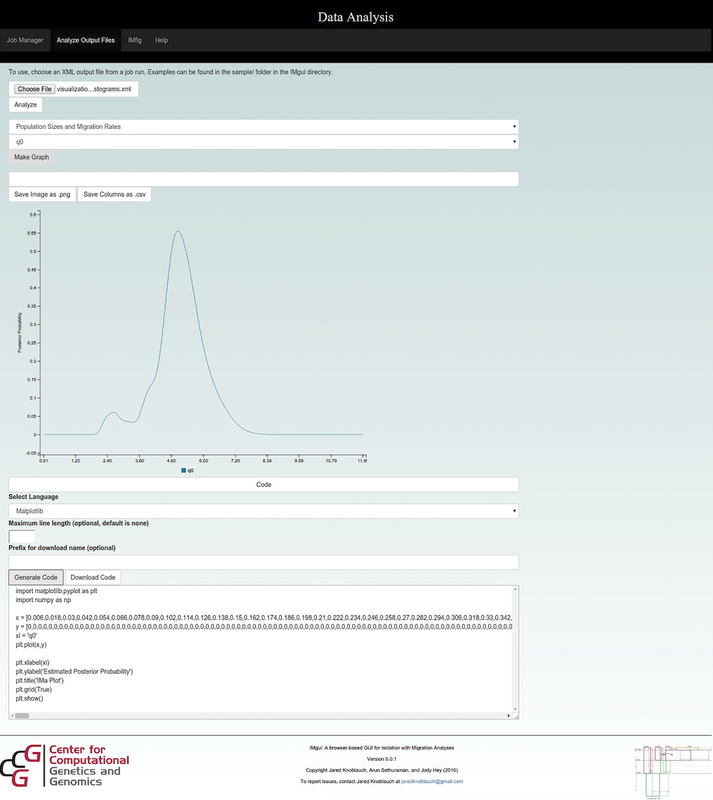
IMGui |
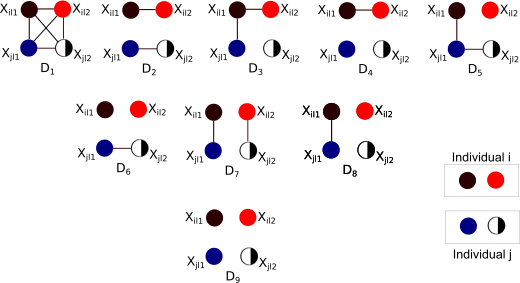
InRelateInRelate is a maximum likelihood method for the estimation of pairwise genetic relatedness between two individuals, under the admixture model. Read more about InRelate in my paper published in G3 here. R code and instructions to use InRelate are here. Khuyen Nguyen has also developed an R package that implements InRelate, which can be accessed here.
|
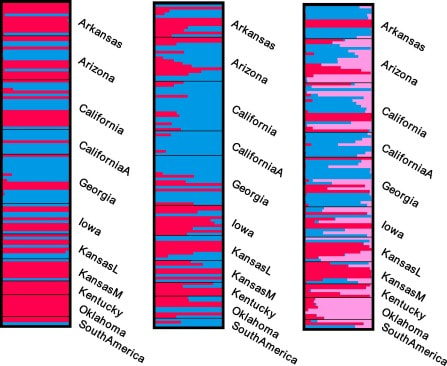
p-MULTICLUSTFast and parallelized estimation of genetic ancestry and allele frequencies from polyploid, multilocus, multi-allelic genomic data under the admixture model. Read more about MULTICLUST here (publication on it is coming soon!).
|
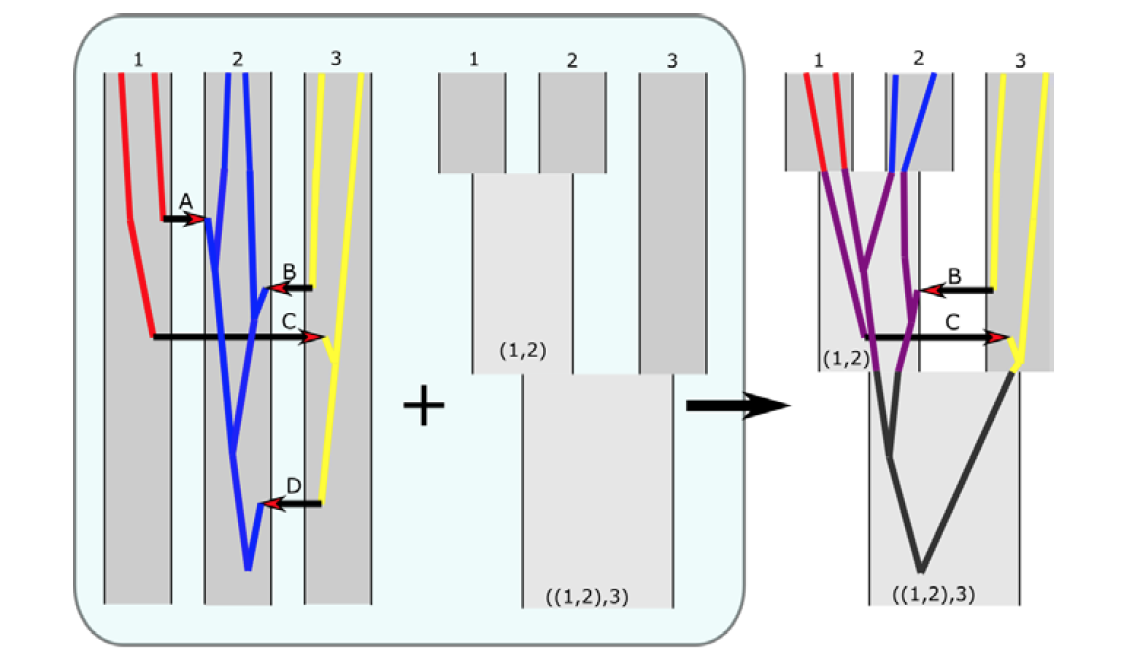
MigSelectParallelized framework for the estimation of genomic loci under the effects of selection against migration (genomic islands of speciation), or those under the effects of linked natural selection effects. Read the methods paper here (publication on the parallelized implementation, and applications to genomic data coming soon, but watch this video from Dr. Sethuraman at the SSE meetings to learn more). You can also access the code implementing MigSelect here.
|
IMa3 |
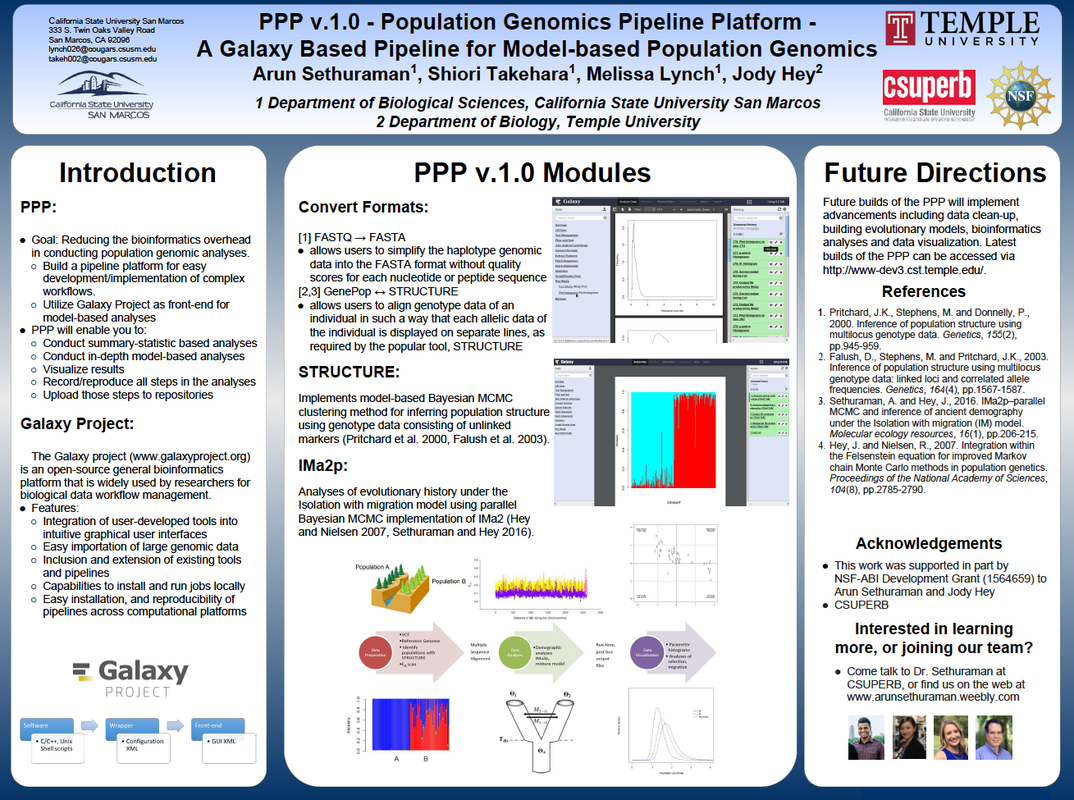
PPP - PopGen Pipeline PlatformAn end-end implementation of scripts for QC, filtering, performing summary statistics estimation, and model-based estimation of evolutionary histories from population genomic data. This project is still in development, but you can track our updates here and install and work with v1 at the PPP development site. A manuscript describing PPP has now been published in MBE.
|
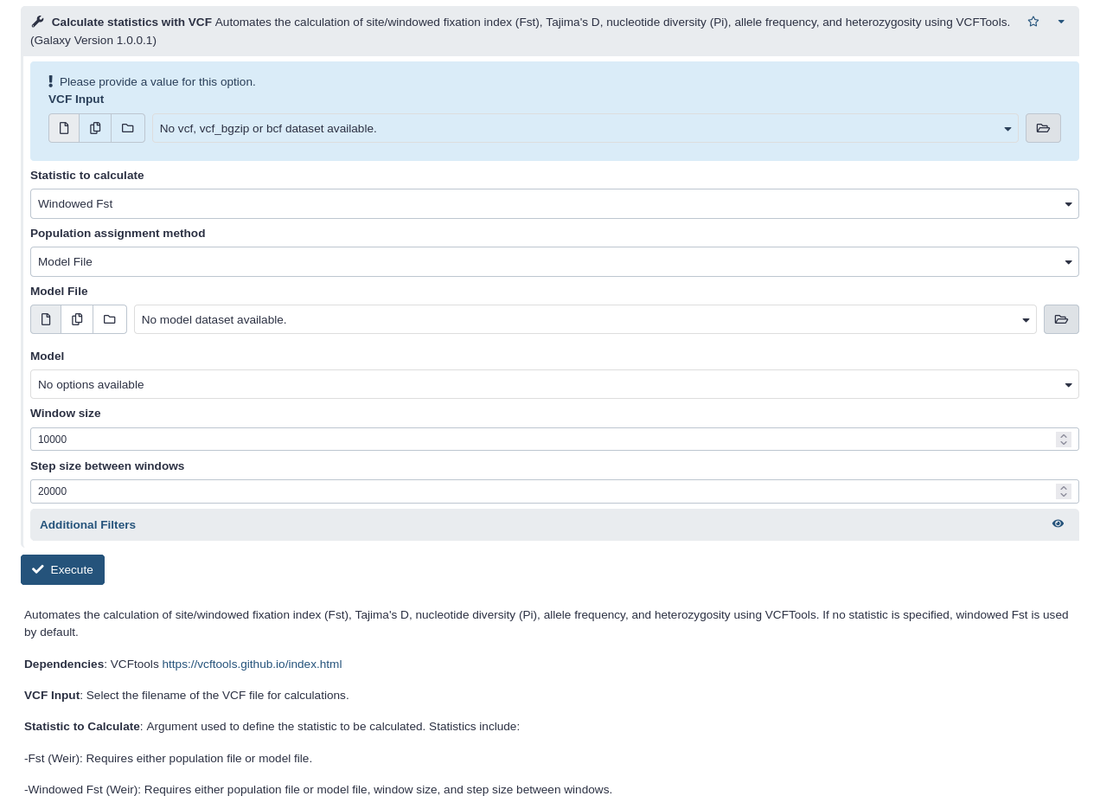
PPPGalaxyPPPGalaxy is an implementation of the PPP (Pop-Gen Pipeline Platform) for automated and reproducible workflows for population genomic analyses on the Galaxy Project Toolshed. PPPGalaxy can be deployed locally or remotely, and provides a web-browser based interface for end users to upload, access, filter, compute a variety of population genomic statistics, perform model-based estimation of population structure and evolutionary demographic history, and data conversions to formats utilized by several popular population genomic methods.
|
ConGenML
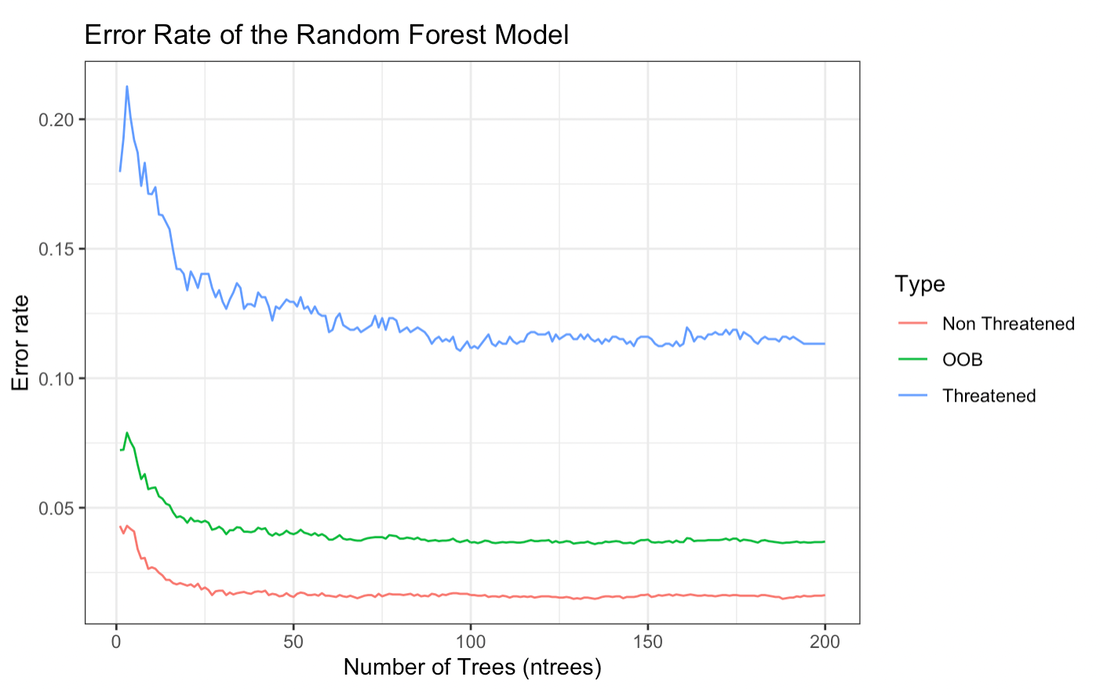
ConGenML implements a deep learning technique to predict IUCN RedList status using a variety of predictors, derived from the MacroPopGen database. This tool was developed by MS student Anais Aoki, and a manuscript describing it can be accessed here.
|
methylMapRChris Morrissey has now developed an R package that combines numerous lines of evidence from long-read sequencing data (PacBio) to characterize the functional methylome in prokaryotic species. Details on how to install and use it are available on his GitHub page here.
|
SpecKsTamsen has now developed a package to simulate whole genome duplications under allo and autopolyploidy, and to estimate their Ks histograms. Watch this space for the manuscript describing this package soon! Meanwhile, we have a preprint describing SpecKs now out here. You can also access SpecKs on Tamsen's Git page here.
|
fscWrapRaya has now developed a wrapper for fastsimcoal28, which permits users to automatically generate and test hundreds of possible models. Watch this space for the manuscript and code pages soon!
|